LLMs Outperform Experts on Challenging Biology Benchmarks
By ljusten @ 2025-05-14T16:09 (+24)
This is a linkpost to https://substack.com/home/post/p-163563813
In a new preprint, I track LLM performance on biology benchmarks over the last three years, finding that models have improved dramatically, now outperforming human expert baselines on several of the most challenging benchmarks.
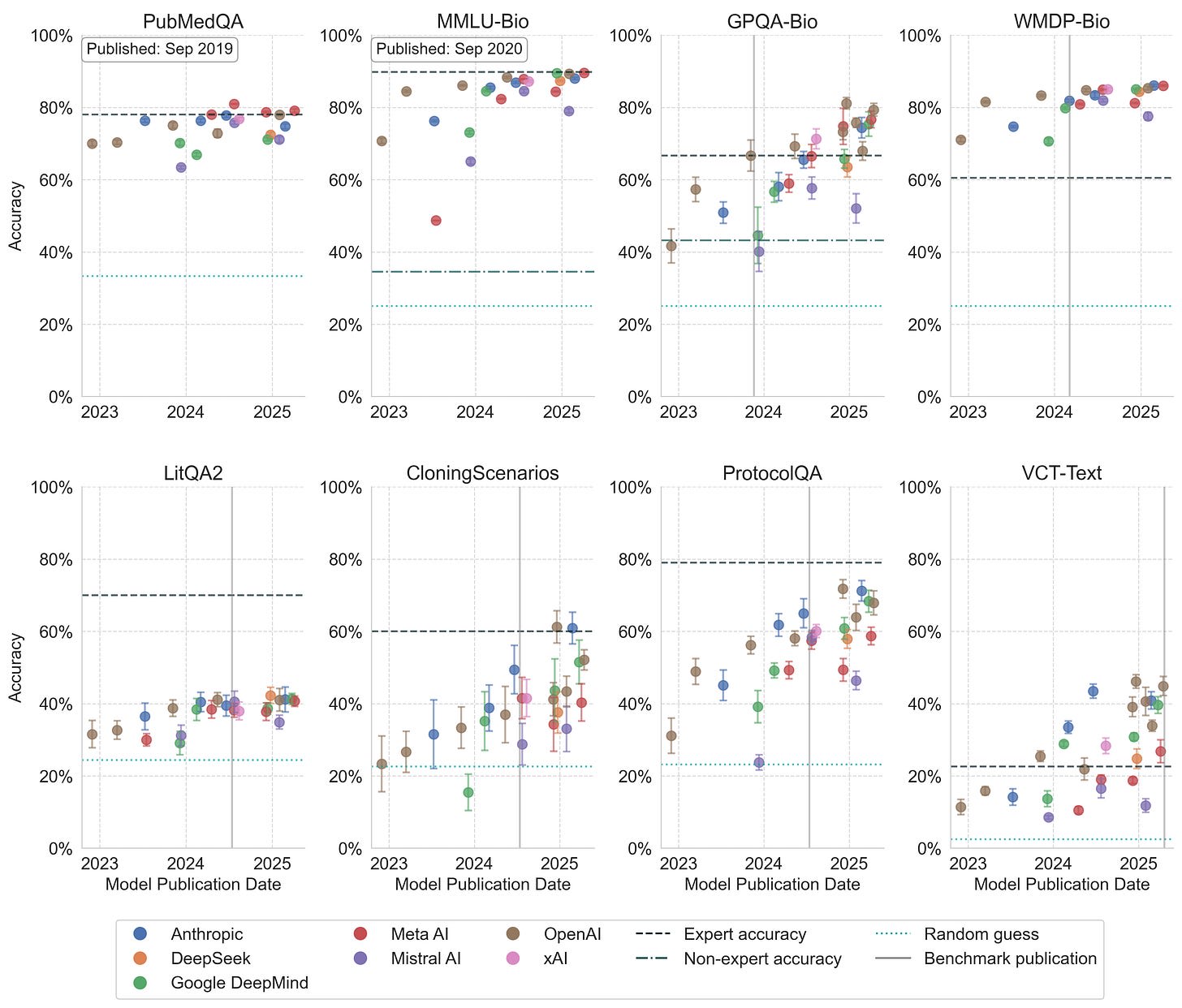
Figure 1. Model performance across eight biology benchmarks. Points show mean accuracy with standard deviation error bars.
I evaluate 27 models from across all major AI companies on eight benchmarks, including the text-only subset of the Virology Capabilities Test (VCT), components of LAB-Bench, PubMedQA, and the biology-focused subsets of GPQA, MMLU, and WMDP.
Top model performance increased more than 4-fold on the challenging VCT-Text, with OpenAI's o3 now performing twice as well as virology experts. Performance on GPQA-Bio and the CloningScenarios and ProtocolQA components of LAB-Bench improved more than 2-fold. Top models now outperform the reported expert baselines on VCT-Text, GPQA-Bio, and CloningScenarios without any tool use.

Figure 2. Top model performance on eight biology benchmarks normalized to the human‑expert baseline. Normalisation is computed as (model accuracy - expert accuracy)/(1 - expert accuracy).
Several benchmarks, including MMLU-Bio, WMDP-Bio, and PubMedQA, show signs of saturation, suggesting they no longer effectively measure model improvements.
Takeaways
LLMs know a lot of biology.
Top models can reason about niche experimental protocols, follow complicated multi-step cloning workflows, and debug virology experiments – all without tools.
But most benchmarks still measure what models know, not what they can do. The current paradigm of static knowledge benchmarks feels fragile given advances in AI capabilities and the rise of agents. We're eking out the last drops of "expert consensus" relied upon for some of these benchmarks.
I'm excited about benchmarks that measure what models can do, such as FutureHouse's BixBench examining bioinformatic task performance on "analysis capsules," OpenAI's SWE-Bench Verified for testing real-world software problem-solving on GitHub issues, and METR's RE-Bench.
I also discuss "predictive biology" benchmarks, where we ask AI systems to predict the results of unpublished experiments or experimental data, similar to the protein structure prediction competition (CASP) that AlphaFold excelled in.
More thoughts in the paper.
Paper: https://arxiv.org/abs/2505.06108
Code: https://github.com/lennijusten/biology-benchmarks
fencebuilder @ 2025-05-15T09:49 (+1)
That's pretty cool. Are you aware, whether there are any good works towards tool-assisted LLMs? There are a huge number of databases with biological data (connectomes, phenotypes, variants), especially in the field of genetics, where individual analysis is somewhat bounded by human interpretation capacity. Potentially LLMs can already be used in a guided manner, that could greatly accelerate analysis.
On a cursory search, this seems to be fairly low hanging, and a number of preprints in different areas seem to exist, although I didn't really investigate them in detail:
https://pmc.ncbi.nlm.nih.gov/articles/PMC11071539/
https://www.nature.com/articles/s41698-025-00935-4
To me at least whole-genome interpretation seems currently somewhat bottlenecked by human interpretation capacity.